This content is from the fall 2016 version of this course. Please go here for the most recent version.
R Markdown, git, and stuff
lab02 - October 5, 2016
Introduction to R Markdown
Console editor vs. scripts vs. R Markdown
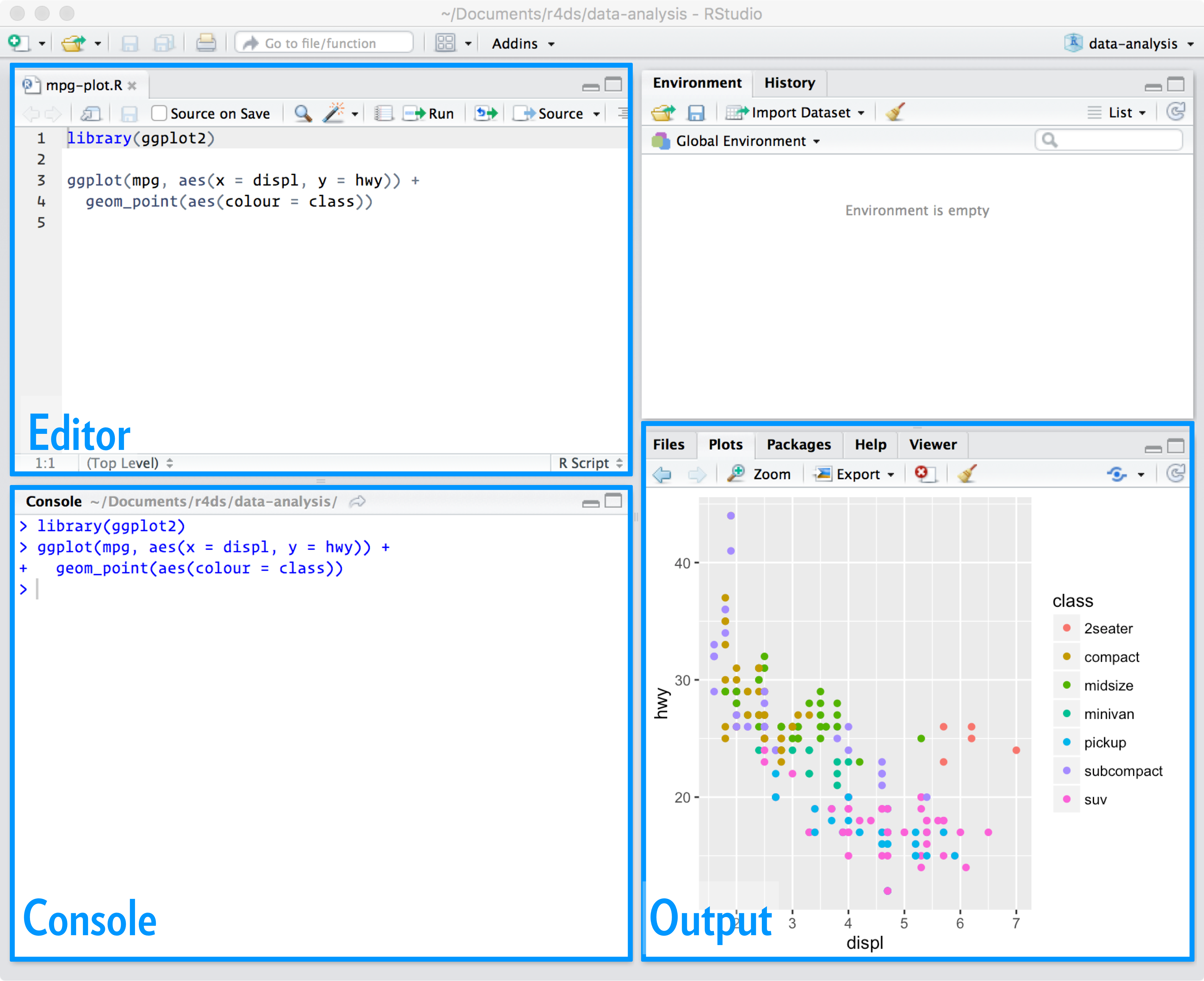
From R for Data Science
- Console editor
- Great for experimenting and interactive coding in R
- No record of past commands
- Must run one line at a time
- Script editor
- Build code in chunks, then run all at once
- Save as a
.Rfile (called an R script) - Can run:
- One line at a time (Cmd/Ctrl + Enter)
- Several lines at once (highlight the code with the cursor, then Cmd/Ctrl + Enter)
- Run the entire script at once (Cmd/Ctrl + Shift + S)
- Output is printed in the console
- Plots are displayed in the bottom-right panel
- Can split a complicated program/workflow into multiple and distinct R scripts (easier to organize large chunks of code)
- R Markdown
- Provides a unified authoring framework for data science
- Combines:
- Code
- Results
- Written commentary
- Displays output and plots within the document (can be changed)
- Good for a final report
- During class, usually better to work in an R script until you are comfortable using R Markdown for homework
Assorted things about Git and GitHub
.gitignore
By default, Git tracks all directories and files in your repository. Sometimes you may not want it to track everything. For instance, if you store a private API key or personally-identifiable data, you won’t want these files tracked by Git. If you did, when you push your repository to GitHub your private files will be shared with the world.
You could just store all of these files outside your repository, but that’s a pain and inconvenient. Instead, you can create a .gitignore file in your repository. This is a special file Git uses to determine what files it should ignore. Any file listed in .gitignore will not be tracked by Git.
When you create a new repository in GitHub (as opposed to forking an existing one), you have the option to add a template .gitignore file depending on what programming language you will use. For example, the default .gitignore file for R is
# History files
.Rhistory
.Rapp.history
# Session Data files
.RData
# Example code in package build process
*-Ex.R
# Output files from R CMD build
/*.tar.gz
# Output files from R CMD check
/*.Rcheck/
# RStudio files
.Rproj.user/
# produced vignettes
vignettes/*.html
vignettes/*.pdf
# OAuth2 token, see https://github.com/hadley/httr/releases/tag/v0.3
.httr-oauth
# knitr and R markdown default cache directories
/*_cache/
/cache/
# Temporary files created by R markdown
*.utf8.md
*.knit.md
.Rproj.userMost of these files are not sensitive, but are merely temporary work files that you don’t need to save and track using version control. You can specify files and directories by their full name, a partial name, or file extension. Starting with homework 2 I will always include a .gitignore in the repository, but for your own projects you will need to create these files as you find necessary.
Clone from the fork, not the master
Make sure whenever you clone a homework repository, use the url for the forked version, not the master repository. So for the first homework, I would use https://github.com/bensoltoff/hw01 when I clone the repo, not https://github.com/uc-cfss/hw01. If you use the master repo url, you will get an error when you try to push your changes to GitHub.
For an example, let’s say I wanted to make a contribution to ggplot2. I should fork the repo and clone the fork. Instead I goofed and cloned the original repo. When I try to push my change, I get an error message:
remote: Permission to hadley/ggplot2.git denied to bensoltoff.
fatal: unable to access 'https://github.com/hadley/ggplot2.git/': The requested URL returned error: 403I don’t have permission to edit the master repo on Hadley Wickham’s account.
How do I fix this? I could go back and clone the correct fork, but if I’ve already made several commits then I’ll lose all my work. Instead, I can change the upstream url: this changes the location Git tries to push my changes. To do this:
- Open up the shell
- Change the current working directory to your local project (should use the
cdcommand) - List your existing remotes in order to get the name of the remote you want to change.
Benjamins-MacBook-Pro:ggplot2 soltoffbc$ git remote -v
origin https://github.com/hadley/ggplot2.git (fetch)
origin https://github.com/hadley/ggplot2.git (push)- Change your remote’s URL to the fork with the
git remote set-urlcommand.
Benjamins-MacBook-Pro:ggplot2 soltoffbc$ git remote set-url origin https://github.com/bensoltoff/ggplot2- Verify that the remote URL has changed.
Benjamins-MacBook-Pro:ggplot2 soltoffbc$ git remote -v
origin https://github.com/bensoltoff/ggplot2 (fetch)
origin https://github.com/bensoltoff/ggplot2 (push)Now I can push successfully to my fork, then submit a pull request.
Use the proper shell (GitBash for Windows)
Make sure to use the proper program when entering the shell. For Mac users, that is Terminal. For Windows users, that is GitBash: if you followed the setup instructions properly, you should have this program on your computer. Look for it under the Start Menu > Git > GitBash. If you try to use the Command Prompt, you will run into errors because it uses different commands than GitBash.
Variable assignment vs. piping
Remember that with pipes, we don’t have to save all of our intermediate steps. We only use one assignment, like this:
(diamonds_summary <- diamonds %>%
filter(carat > .2, carat < 2) %>%
group_by(cut, color) %>%
summarize(price = mean(price, na.rm = TRUE),
depth = mean(depth, na.rm = TRUE))
)## Source: local data frame [35 x 4]
## Groups: cut [?]
##
## cut color price depth
## <ord> <ord> <dbl> <dbl>
## 1 Fair D 3865.121 64.02866
## 2 Fair E 3406.472 63.29174
## 3 Fair F 3441.201 63.48188
## 4 Fair G 3331.885 64.19928
## 5 Fair H 3922.667 64.53566
## 6 Fair I 3516.860 64.05733
## 7 Fair J 3323.617 64.10638
## 8 Good D 3234.587 62.36570
## 9 Good E 3246.772 62.22587
## 10 Good F 3286.783 62.19631
## # ... with 25 more rowsDo not do this:
(diamonds_summary <- diamonds %>%
diamonds_filter <- filter(carat > .2, carat < 2) %>%
diamonds_group <- group_by(cut, color) %>%
diamonds_summary <- summarize(price = mean(price, na.rm = TRUE),
depth = mean(depth, na.rm = TRUE))
)## Error in summarise_(.data, .dots = lazyeval::lazy_dots(...)): argument ".data" is missing, with no defaultOr this:
(diamonds_summary <- diamonds %>%
filter(diamonds, carat > .2, carat < 2) %>%
group_by(diamonds, cut, color) %>%
summarize(diamonds,
price = mean(price, na.rm = TRUE),
depth = mean(depth, na.rm = TRUE))
)## Warning in Ops.ordered(left, right): '&' is not meaningful for ordered
## factors
## Warning in Ops.ordered(left, right): '&' is not meaningful for ordered
## factors
## Warning in Ops.ordered(left, right): '&' is not meaningful for ordered
## factors## Error in eval(expr, envir, enclos): incorrect length (539400), expecting: 53940If you use pipes, you don’t have to call the data frame with each function - just the first time.
Session Info
Session information:
devtools::session_info()## Session info --------------------------------------------------------------## setting value
## version R version 3.3.1 (2016-06-21)
## system x86_64, darwin13.4.0
## ui RStudio (1.0.44)
## language (EN)
## collate en_US.UTF-8
## tz America/Chicago
## date 2016-11-16## Packages ------------------------------------------------------------------## package * version date source
## assertthat 0.1 2013-12-06 CRAN (R 3.3.0)
## codetools 0.2-15 2016-10-05 CRAN (R 3.3.0)
## colorspace 1.2-7 2016-10-11 CRAN (R 3.3.0)
## DBI 0.5-1 2016-09-10 CRAN (R 3.3.0)
## devtools 1.12.0 2016-06-24 CRAN (R 3.3.0)
## digest 0.6.10 2016-08-02 CRAN (R 3.3.0)
## dplyr * 0.5.0 2016-06-24 CRAN (R 3.3.0)
## evaluate 0.10 2016-10-11 CRAN (R 3.3.0)
## formatR 1.4 2016-05-09 CRAN (R 3.3.0)
## gapminder * 0.2.0 2015-12-31 CRAN (R 3.3.0)
## ggplot2 * 2.2.0 2016-11-10 Github (hadley/ggplot2@f442f32)
## gtable 0.2.0 2016-02-26 CRAN (R 3.3.0)
## htmltools 0.3.5 2016-03-21 CRAN (R 3.3.0)
## knitr 1.15 2016-11-09 CRAN (R 3.3.1)
## labeling 0.3 2014-08-23 CRAN (R 3.3.0)
## lattice 0.20-34 2016-09-06 CRAN (R 3.3.0)
## lazyeval 0.2.0 2016-06-12 CRAN (R 3.3.0)
## lubridate * 1.6.0 2016-09-13 CRAN (R 3.3.0)
## magrittr 1.5 2014-11-22 CRAN (R 3.3.0)
## Matrix 1.2-7.1 2016-09-01 CRAN (R 3.3.0)
## memoise 1.0.0 2016-01-29 CRAN (R 3.3.0)
## mgcv 1.8-16 2016-11-07 CRAN (R 3.3.0)
## munsell 0.4.3 2016-02-13 CRAN (R 3.3.0)
## nlme 3.1-128 2016-05-10 CRAN (R 3.3.1)
## plyr 1.8.4 2016-06-08 CRAN (R 3.3.0)
## purrr * 0.2.2 2016-06-18 CRAN (R 3.3.0)
## R6 2.2.0 2016-10-05 CRAN (R 3.3.0)
## randomForest 4.6-12 2015-10-07 CRAN (R 3.3.0)
## rcfss * 0.1.0 2016-10-06 local
## Rcpp 0.12.7 2016-09-05 cran (@0.12.7)
## readr * 1.0.0 2016-08-03 CRAN (R 3.3.0)
## readxl * 0.1.1 2016-03-28 CRAN (R 3.3.0)
## rmarkdown * 1.1 2016-10-16 CRAN (R 3.3.1)
## rsconnect 0.5 2016-10-17 CRAN (R 3.3.0)
## rstudioapi 0.6 2016-06-27 CRAN (R 3.3.0)
## scales 0.4.1 2016-11-09 CRAN (R 3.3.1)
## stringi 1.1.2 2016-10-01 CRAN (R 3.3.0)
## stringr * 1.1.0 2016-08-19 cran (@1.1.0)
## tibble * 1.2 2016-08-26 cran (@1.2)
## tidyr * 0.6.0 2016-08-12 CRAN (R 3.3.0)
## tidyverse * 1.0.0 2016-09-09 CRAN (R 3.3.0)
## withr 1.0.2 2016-06-20 CRAN (R 3.3.0)
## yaml 2.1.13 2014-06-12 CRAN (R 3.3.0)This work is licensed under the CC BY-NC 4.0 Creative Commons License.